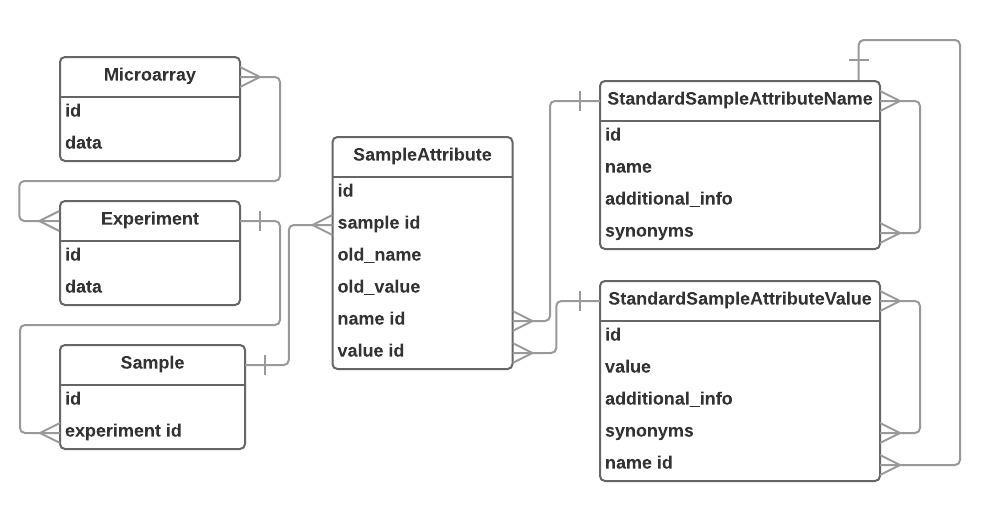
The database used by the portal was designed to accommodate varying amounts of sample attributes, ensure reverse compatibility with original metadata, provide standardized names (from MeSH and EFO) in sample annotations, and be extendable for including data on other -omics (genome, metabolome, transcriptome, etc.) and obtained using different platforms (NGS, for example). The "Database Structure" diagram represents the relations between the tables in the database. More details are available in our article by Lykhenko, O., Frolova, A., & Obolenskaya, M.(2017). Creation of gene expression database on preeclampsia-affected human placenta. Biopolym. Cell, 33(6), 442–452. doi: 10.7124/bc.000967.